主要信息
Target
E2F-4
Host Species
Rabbit
Reactivity
Human, Mouse, Rat
Applications
WB, ELISA
MW
43kD (Observed)
Conjugate/Modification
Acetyl
货号: YK0025
规格
价格
货期
数量
200μL
¥4,680.00
现货
0
100μL
¥2,800.00
现货
0
50μL
¥1,500.00
现货
0
加入购物车


已收藏


收藏
详细信息
推荐稀释比
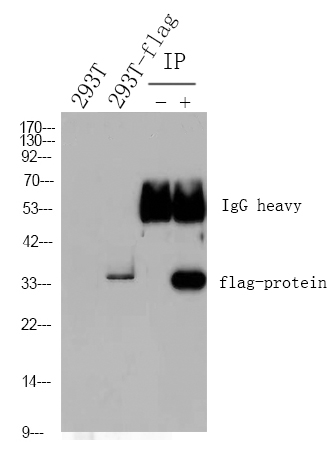
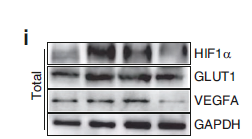
WB 1:500-1:2000; ELISA 1:20000; Not yet tested in other applications.
组成
Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
特异性
Acetyl-E2F-4 (K96) Polyclonal Antibody detects endogenous levels of E2F-4 protein only when acetylated at K96.The name of modified sites may be influenced by many factors, such as species (the modified site was not originally found in human samples) and the change of protein sequence (the previous protein sequence is incomplete, and the protein sequence may be prolonged with the development of protein sequencing technology). When naming, we will use the "numbers" in historical reference to keep the sites consistent with the reports. The antibody binds to the following modification sequence (lowercase letters are modification sites):ADkLI
纯化工艺
The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
储存
-15°C to -25°C/1 year(Do not lower than -25°C)
浓度
1 mg/ml
实测条带
43kD
修饰
Acetyl
克隆性
Polyclonal
同种型
IgG
相关产品
抗原&靶点信息
免疫原:
The antiserum was produced against synthesized Acetyl-peptide derived from human E2F4 around the Acetylation site of Lys96. AA range:61-110
展开内容
特异性:
Acetyl-E2F-4 (K96) Polyclonal Antibody detects endogenous levels of E2F-4 protein only when acetylated at K96.The name of modified sites may be influenced by many factors, such as species (the modified site was not originally found in human samples) and the change of protein sequence (the previous protein sequence is incomplete, and the protein sequence may be prolonged with the development of protein sequencing technology). When naming, we will use the "numbers" in historical reference to keep the sites consistent with the reports. The antibody binds to the following modification sequence (lowercase letters are modification sites):ADkLI
展开内容
基因名称:
E2F4
展开内容
蛋白名称:
Transcription factor E2F4
展开内容
别名:
E2F4 ;
Transcription factor E2F4 ;
E2F-4
Transcription factor E2F4 ;
E2F-4
展开内容
背景:
The protein encoded by this gene is a member of the E2F family of transcription factors. The E2F family plays a crucial role in the control of cell cycle and action of tumor suppressor proteins and is also a target of the transforming proteins of small DNA tumor viruses. The E2F proteins contain several evolutionally conserved domains found in most members of the family. These domains include a DNA binding domain, a dimerization domain which determines interaction with the differentiation regulated transcription factor proteins (DP), a transactivation domain enriched in acidic amino acids, and a tumor suppressor protein association domain which is embedded within the transactivation domain. This protein binds to all three of the tumor suppressor proteins pRB, p107 and p130, but with higher affinity to the last two. It plays an important role in the suppression of proliferation-associated ge
展开内容
功能:
developmental stage:Present in the growth-arrested state, its abundance does not change significantly as cells move into and through the cell cycle.,Function:Transcription activator that binds DNA cooperatively with DP proteins through the E2 recognition site, 5'-TTTC[CG]CGC-3' found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA replication. The DRTF1/E2F complex functions in the control of cell-cycle progression from G1 to S phase. E2F-4 binds with high affinity to RBL1 and RBL2. In some instances, can also bind RB protein.,polymorphism:The poly-Ser region of E2F4 is polymorphic and the number of Ser varies in the population (from 8 to 17). The variation might be associated with tumorigenesis.,PTM:Differentially phosphorylated in vivo.,similarity:Belongs to the E2F/DP family.,subunit:Component of the DRTF1/E2F transcription factor complex. Binds cooperatively with DP-1 to E2F sites. The E2F4/DP-1 dimer interacts preferentially with pocket protein RBL1, which inhibits the E2F transactivation domain. Lower affinity interaction has been found with retinoblastoma protein RB1. Interacts with TRRAP, which probably mediates its interaction with histone acetyltransferase complexes, leading to transcription activation. Interacts with HCFC1. Component of the DREAM complex (also named LINC complex) at least composed of E2F4, E2F5, LIN9, LIN37, LIN52, LIN54, MYBL1, MYBL2, RBL1, RBL2, RBBP4, TFDP1 and TFDP2. The complex exists in quiescent cells where it represses cell cycle-dependent genes. It dissociates in S phase when LIN9, LIN37, LIN52 and LIN54 form a subcomplex that binds to MYBL2.,tissue specificity:Found in all tissue examined including heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas.,
展开内容
细胞定位:
Nucleus.
展开内容
组织表达:
Found in all tissue examined including heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas.
展开内容
研究领域:
>>Cell cycle ;
>>Cellular senescence ;
>>TGF-beta signaling pathway
>>Cellular senescence ;
>>TGF-beta signaling pathway
展开内容
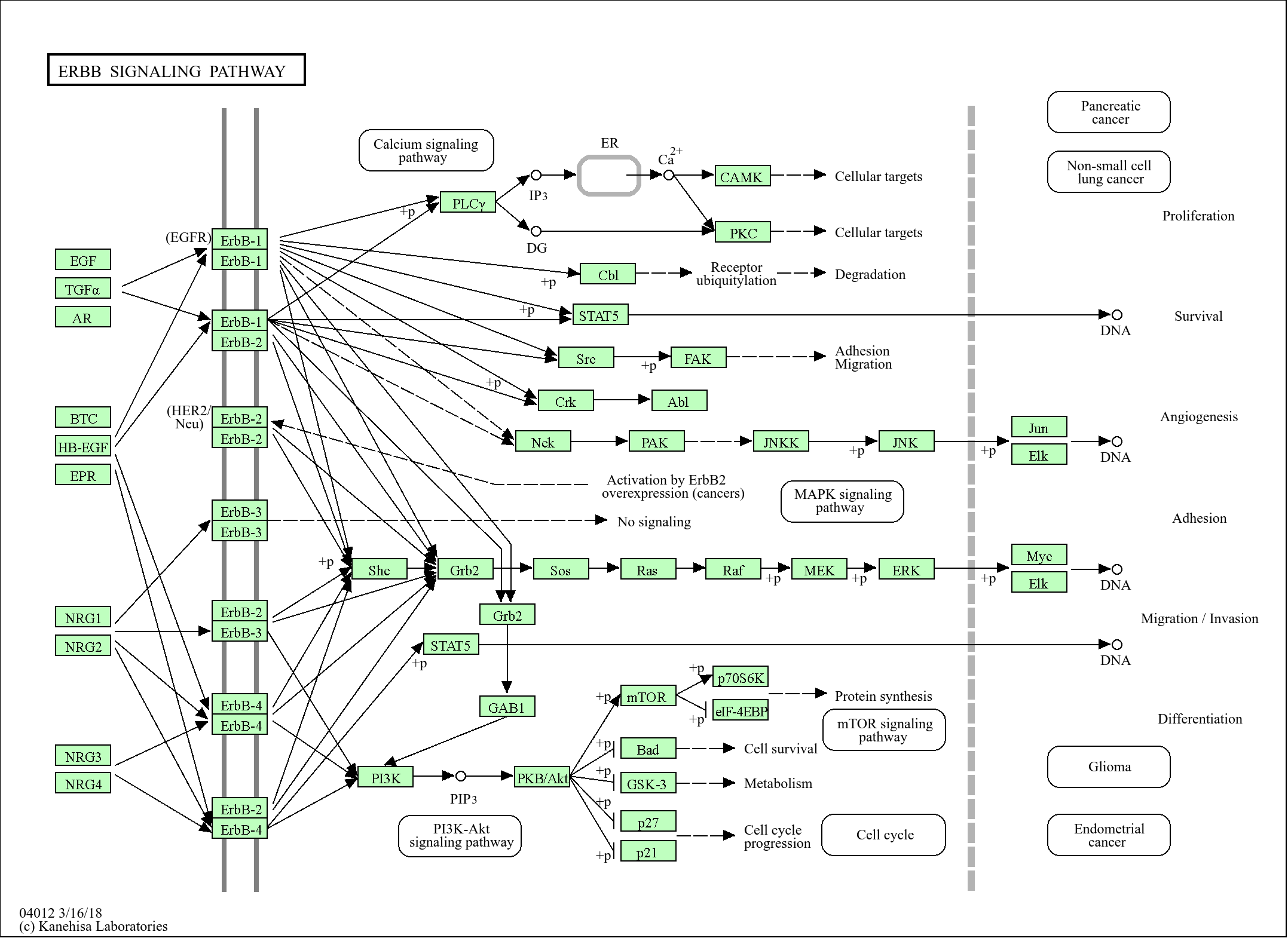
信号通路
文献引用({{totalcount}})
货号: YK0025
规格
价格
货期
数量
200μL
¥4,680.00
现货
0
100μL
¥2,800.00
现货
0
50μL
¥1,500.00
现货
0
加入购物车


已收藏


收藏
Recently Viewed Products
Clear allToggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
主要信息
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
产品 {{index}}/{{pcount}}
上一个产品
下一个产品
{{pvTitle}}
滚轮缩放图片
{{pvDescr}}